 京公网安备 11010802034615号
经营许可证编号:京B2-20210330
京公网安备 11010802034615号
经营许可证编号:京B2-20210330
对于有SQL背景的R语言学习者而言,sqldf是一个非常有用的包,因为它使我们能在R中使用SQL命令。只要掌握了基本的SQL技术,我们就能利用它们在R中操作数据框。关于sqldf包的更多信息,可以参看 cran 。
在这篇文章中,我们将展示如何在R中利用SQL命令来连接、检索、排序和筛选数据。我们也将展示怎么利用R语言的函数来实现这些功能。最近我在处理一些FDA(译者注:食品及药物管理局)的不良事件数据。这些数据非常混乱:有缺失值,有重复记录,有不同时间建立的数据集的可比性问题,不同数据集中变量名称和数量也不统一(比如一个数据集里叫sex,另一个里叫gender),还有疏忽错误等问题。但正因如此,这些数据对于数据科学家或者爱好者而言到是理想的练手对象。
本文使用的FDA不良事件数据可以从公开渠道获得,csv格式的数据表可以从国家经济研究局下载。通过R从国家经济研究局的网站下载数据相对更容易,我建议你使用相应的R代码来下载并探索数据。
不良事件数据集是以季度为发布周期,每个季度的数据包括了人口信息、药物/生物信息、不良事件详情,结果和诊断情况等信息。
让我们下载数据并使用SQL命令来连接、排序和筛选该数据集中包含的大量数据框。
加载R包
require(downloader)
library(dplyr)
library(sqldf)
library(data.table)
library(ggplot2)
library(compare)
library(plotrix)
基本的错误处理函数tryCatch()
我们将使用这个函数来处理下载的数据。因为数据以季度频率发布,每年都会有四个观测值(每年有四条记录)。运行这个函数能自动下载数据,但如果某些季度数据从网上无法获取(尚未公布),该函数会返回一条错误信息表示无法找到数据集。现在让我们下载数据的压缩包并将其解压。
try.error = function(url)
{
try_error = tryCatch(download(url,dest="data.zip"), error=function(e) e)
if (!inherits(try_error, "error")){
download(url,dest="data.zip")
unzip ("data.zip")
}
else if (inherits(try_error, "error")){
cat(url,"not found\n")
}
}
下载不良事件数据
我们可以得到自2004年起的FDA不良事件数据。本文将使用2013年以来公布的数据,我们将检查截至当前时间的最新数据并下载。
> Sys.time() 函数会返回当前的日期和时间。数据分析师培训
> data.table包中的year()函数会从之前返回的当前时间中提取年份信息。
我们将下载人口、药物、诊断/指示,结果和反应(不良事件)数据。
year_start=2013
year_last=year(Sys.time())
for (i in year_start:year_last){
j=c(1:4)
for (m in j){
url1<-paste0("http://www.nber.org/fda/faers/",i,"/demo",i,"q",m,".csv.zip")
url2<-paste0("http://www.nber.org/fda/faers/",i,"/drug",i,"q",m,".csv.zip")
url3<-paste0("http://www.nber.org/fda/faers/",i,"/reac",i,"q",m,".csv.zip")
url4<-paste0("http://www.nber.org/fda/faers/",i,"/outc",i,"q",m,".csv.zip")
url5<-paste0("http://www.nber.org/fda/faers/",i,"/indi",i,"q",m,".csv.zip")
try.error(url1)
try.error(url2)
try.error(url3)
try.error(url4)
try.error(url5)
}
}
http://www.nber.org/fda/faers/2015/demo2015q4.csv.zip not found
...
http://www.nber.org/fda/faers/2016/indi2016q4.csv.zip not found
根据上面的错误信息,截至成文时间(2016年3月13日),我们最多可以获得2015年第三季度的不良事件数据。
> list.files()函数会字符串向量的形式返回当前工作目录下所有文件的名字。
> 我会使用正则表达式对各个数据集的类别进行筛选。比如^demo.*.csv表示所有名字以demo开头的csv文件。
filenames <- list.files(pattern="^demo.*.csv", full.names=TRUE)
cat('We have downloaded the following quarterly demography datasets')
filenames
我们已经下载了下列季度人口数据
"./demo2012q1.csv" "./demo2012q2.csv" "./demo2012q3.csv" "./demo2012q4.csv" "./demo2013q1.csv" "./demo2013q2.csv" "./demo2013q3.csv" "./demo2013q4.csv" "./demo2014q1.csv" "./demo2014q2.csv" "./demo2014q3.csv" "./demo2014q4.csv" "./demo2015q1.csv" "./demo2015q2.csv" "./demo2015q3.csv"
让我们用data.table包中的fread()函数来读入这些数据集,以人口数据为例:
demo=lapply(filenames,fread)
接着让我们把它们转换数据结构并合并成一个数据框:
demo_all=do.call(rbind,lapply(1:length(demo),function(i) select(as.data.frame(demo[i]),primaryid,caseid, age,age_cod,event_dt,sex,reporter_country)))
dim(demo_all)
3554979 7
我们看到人口数据有超过350万行观测(记录)。
译者注:下面的内容都是重复这个流程,可以略过
现在让我们合并所有的药品数据
filenames <- list.files(pattern="^drug.*.csv", full.names=TRUE)
cat('We have downloaded the following quarterly drug datasets:\n')
filenames
drug=lapply(filenames,fread)
cat('\n')
cat('Variable names:\n')
names(drug[[1]])
drug_all=do.call(rbind,lapply(1:length(drug), function(i) select(as.data.frame(drug[i]),primaryid,caseid, drug_seq,drugname,route)))
我们已经下载了下列季度药品数据集
"./drug2012q1.csv" "./drug2012q2.csv" "./drug2012q3.csv" "./drug2012q4.csv" "./drug2013q1.csv" "./drug2013q2.csv" "./drug2013q3.csv" "./drug2013q4.csv" "./drug2014q1.csv" "./drug2014q2.csv" "./drug2014q3.csv" "./drug2014q4.csv" "./drug2015q1.csv" "./drug2015q2.csv" "./drug2015q3.csv"
每张表中的变量名分别为:
"primaryid" "drug_seq" "role_cod" "drugname" "val_vbm" "route" "dose_vbm" "dechal" "rechal" "lot_num" "exp_dt" "exp_dt_num" "nda_num"
合并所有的诊断/指示数据集
filenames <- list.files(pattern="^indi.*.csv", full.names=TRUE)
cat('We have downloaded the following quarterly diagnoses/indications datasets:\n')
filenames
indi=lapply(filenames,fread)
cat('\n')
cat('Variable names:\n')
names(indi[[15]])
indi_all=do.call(rbind,lapply(1:length(indi), function(i) select(as.data.frame(indi[i]),primaryid,caseid, indi_drug_seq,indi_pt)))
已经下载的数据集为:
"./indi2012q1.csv" "./indi2012q2.csv" "./indi2012q3.csv" "./indi2012q4.csv" "./indi2013q1.csv" "./indi2013q2.csv" "./indi2013q3.csv" "./indi2013q4.csv" "./indi2014q1.csv" "./indi2014q2.csv" "./indi2014q3.csv" "./indi2014q4.csv" "./indi2015q1.csv" "./indi2015q2.csv" "./indi2015q3.csv"
变量名为:
"primaryid" "caseid" "indi_drug_seq" "indi_pt"
合并病人的结果数据:
filenames <- list.files(pattern="^outc.*.csv", full.names=TRUE)
cat('We have downloaded the following quarterly patient outcome datasets:\n')
filenames
outc_all=lapply(filenames,fread)
cat('\n')
cat('Variable names\n')
names(outc_all[[1]])
names(outc_all[[4]])
colnames(outc_all[[4]])=c("primaryid", "caseid", "outc_cod")
outc_all=do.call(rbind,lapply(1:length(outc_all), function(i) select(as.data.frame(outc_all[i]),primaryid,outc_cod)))
下载的数据集如下:
"./outc2012q1.csv" "./outc2012q2.csv" "./outc2012q3.csv" "./outc2012q4.csv" "./outc2013q1.csv" "./outc2013q2.csv" "./outc2013q3.csv" "./outc2013q4.csv" "./outc2014q1.csv" "./outc2014q2.csv" "./outc2014q3.csv" "./outc2014q4.csv" "./outc2015q1.csv" "./outc2015q2.csv" "./outc2015q3.csv"
变量名:
"primaryid" "outc_cod"
"primaryid" "caseid" "outc_code"
最后来合并反应(不良事件)数据集(译者注:这部分无聊地我要哭了)
filenames <- list.files(pattern="^reac.*.csv", full.names=TRUE)
cat('We have downloaded the following quarterly reaction (adverse event) datasets:\n')
filenames
reac=lapply(filenames,fread)
cat('\n')
cat('Variable names:\n')
names(reac[[3]])
reac_all=do.call(rbind,lapply(1:length(indi), function(i) select(as.data.frame(reac[i]),primaryid,pt)))
下载的数据集有:
"./reac2012q1.csv" "./reac2012q2.csv" "./reac2012q3.csv" "./reac2012q4.csv" "./reac2013q1.csv" "./reac2013q2.csv" "./reac2013q3.csv" "./reac2013q4.csv" "./reac2014q1.csv" "./reac2014q2.csv" "./reac2014q3.csv" "./reac2014q4.csv" "./reac2015q1.csv" "./reac2015q2.csv" "./reac2015q3.csv"
变量名为:
"primaryid" "pt"
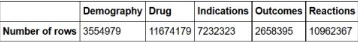
让我们看看不同的数据类型各有多少行
all=as.data.frame(list(Demography=nrow(demo_all),Drug=nrow(drug_all),
Indications=nrow(indi_all),Outcomes=nrow(outc_all),
Reactions=nrow(reac_all)))
row.names(all)='Number of rows'
all
SQL命令=
记住sqldf包使用SQLite
COUNT
# SQL版本 sqldf("SELECT COUNT(primaryid)as 'Number of rows of Demography data' FROM demo_all;")

# R版本
nrow(demo_all)
3554979
LIMIT命令(显示前几行)
# SQL版本
sqldf("SELECT *
FROM demo_all
LIMIT 6;")

# R版本 head(demo_all,6)

R1=head(demo_all,6)
SQL1 =sqldf("SELECT *
FROM demo_all
LIMIT 6;")
all.equal(R1,SQL1)
TRUE
*译者注:这部分代码验证了SQL命令和R代码的等价性,下同。
WHERE命令
SQL2=sqldf("SELECT * FROM demo_all WHERE sex ='F';")
R2 = filter(demo_all, sex=="F")
identical(SQL2, R2)
TRUE
SQL3=sqldf("SELECT * FROM demo_all WHERE age BETWEEN 20 AND 25;")
R3 = filter(demo_all, age >= 20 & age <= 25)
identical(SQL3, R3)
TRUE
GROUP BY 和 ORDER BY
# SQL版本
sqldf("SELECT sex, COUNT(primaryid) as Total
FROM demo_all
WHERE sex IN ('F','M','NS','UNK')
GROUP BY sex
ORDER BY Total DESC ;")

# R版本
demo_all %>% filter(sex %in%c('F','M','NS','UNK')) %>% group_by(sex) %>%
summarise(Total = n()) %>% arrange(desc(Total))

SQL3 = sqldf("SELECT sex, COUNT(primaryid) as Total
FROM demo_all
GROUP BY sex
ORDER BY Total DESC ;")
R3 = demo_all%>%group_by(sex) %>%
summarise(Total = n())%>%arrange(desc(Total))
compare(SQL3,R3, allowAll=TRUE)
TRUE
dropped attributes
SQL=sqldf("SELECT sex, COUNT(primaryid) as Total
FROM demo_all
WHERE sex IN ('F','M','NS','UNK')
GROUP BY sex
ORDER BY Total DESC ;")
SQL$Total=as.numeric(SQL$Total
pie3D(SQL$Total, labels = SQL$sex,explode=0.1,col=rainbow(4),
main="Pie Chart of adverse event reports by gender",cex.lab=0.5, cex.axis=0.5, cex.main=1,labelcex=1)
输出的图如下:

Inner Join
让我们把药品数据和指数数据基于主id和药品序列内连。
首先,我们要检查下变量名,看看如何合并两个数据集。
names(indi_all)
names(drug_all)
"primaryid" "indi_drug_seq" "indi_pt"
"primaryid" "drug_seq" "drugname" "route"
names(indi_all)=c("primaryid", "drug_seq", "indi_pt" ) # 使两个数据集变量名一致
R4= merge(drug_all,indi_all, by = intersect(names(drug_all), names(indi_all))) # R版本合并
R4=arrange(R3, primaryid,drug_seq,drugname,indi_pt) # R版本排序
SQL4= sqldf("SELECT d.primaryid as primaryid, d.drug_seq as drug_seq, d.drugname as drugname,
d.route as route,i.indi_pt as indi_pt
FROM drug_all d
INNER JOIN indi_all i
ON d.primaryid= i.primaryid AND d.drug_seq=i.drug_seq
ORDER BY primaryid,drug_seq,drugname, i.indi_pt") # SQL版本
compare(R4,SQL4,allowAll=TRUE)
TRUE # 两种方法等价
R5 = merge(reac_all,outc_all,by=intersect(names(reac_all), names(outc_all)))
SQL5 =reac_outc_new4=sqldf("SELECT r.*, o.outc_cod as outc_cod
FROM reac_all r
INNER JOIN outc_all o
ON r.primaryid=o.primaryid
ORDER BY r.primaryid,r.pt,o.outc_cod")
compare(R5,SQL5,allowAll = TRUE)
TRUE
# 绘制不同性别的年龄概率分布密度图
ggplot(sqldf('SELECT age, sex
FROM demo_all
WHERE age between 0 AND 100 AND sex IN ("F","M")
LIMIT 10000;'), aes(x=age, fill = sex))+ geom_density(alpha = 0.6)
绘制出的图如下:

绘制不同结果的年龄年龄概率分布密度图(译者注:后面都是结果的可视化,可略过。原作者的耐心真好。。。)
ggplot(sqldf("SELECT d.age as age, o.outc_cod as outcome
FROM demo_all d
INNER JOIN outc_all o
ON d.primaryid=o.primaryid
WHERE d.age BETWEEN 20 AND 100
LIMIT 20000;"),aes(x=age, fill = outcome))+ geom_density(alpha = 0.6)
输出如下:

ggplot(sqldf("SELECT de.sex as sex, dr.route as route
FROM demo_all de
INNER JOIN drug_all dr
ON de.primaryid=dr.primaryid
WHERE de.sex IN ('M','F') AND dr.route IN ('ORAL','INTRAVENOUS','TOPICAL')
LIMIT 200000;"),aes(x=route, fill = sex))+ geom_bar(alpha=0.6)
输出如下:

ggplot(sqldf("SELECT d.sex as sex, o.outc_cod as outcome
FROM demo_all d
INNER JOIN outc_all o
ON d.primaryid=o.primaryid
WHERE d.age BETWEEN 20 AND 100 AND sex IN ('F','M')
LIMIT 20000;"),aes(x=outcome,fill=sex))+ geom_bar(alpha = 0.6)
输出如下(译者注:哥们儿挺住,你就快看完了!!!):

UNION ALL
demo1= demo_all[1:20000,]
demo2=demo_all[20001:40000,]
R6 <- rbind(demo1, demo2)
SQL6 <- sqldf("SELECT * FROM demo1 UNION ALL SELECT * FROM demo2;")
compare(R6,SQL6, allowAll = TRUE)
TRUE
INTERSECT
R7 <- semi_join(demo1, demo2)
SQL7 <- sqldf("SELECT * FROM demo1 INTERSECT SELECT * FROM demo2;")
compare(R7,SQL7, allowAll = TRUE)
TRUE
EXCEPT
R8 <- anti_join(demo1, demo2)
SQL8 <- sqldf("SELECT * FROM demo1 EXCEPT SELECT * FROM demo2;")
compare(R8,SQL8, allowAll = TRUE)
TRUE
翻译感悟:这篇文章的作者不厌其烦地演示了利用如何sqldf包在R中实现大部分常用的SQL命令,并将其结果和直接调用相应的R函数的结果做了对照,证明了二者的等价性。

数据分析咨询请扫描二维码
若不方便扫码,搜微信号:CDAshujufenxi
CDA一级知识点汇总手册 第四章 战略与业务数据分析考点43:战略数据分析基础考点44:表格结构数据的使用考点45:输入数据和资源 ...
2026-02-22CDA一级知识点汇总手册 第三章 商业数据分析框架考点27:商业数据分析体系的核心逻辑——BSC五视角框架考点28:战略视角考点29: ...
2026-02-20CDA一级知识点汇总手册 第二章 数据分析方法考点7:基础范式的核心逻辑(本体论与流程化)考点8:分类分析(本体论核心应用)考 ...
2026-02-18第一章:数据分析思维考点1:UVCA时代的特点考点2:数据分析背后的逻辑思维方法论考点3:流程化企业的数据分析需求考点4:企业数 ...
2026-02-16在数据分析、业务决策、科学研究等领域,统计模型是连接原始数据与业务价值的核心工具——它通过对数据的规律提炼、变量关联分析 ...
2026-02-14在SQL查询实操中,SELECT * 与 SELECT 字段1, 字段2,...(指定个别字段)是最常用的两种查询方式。很多开发者在日常开发中,为了 ...
2026-02-14对CDA(Certified Data Analyst)数据分析师而言,数据分析的核心不是孤立解读单个指标数值,而是构建一套科学、完整、贴合业务 ...
2026-02-14在Power BI实操中,函数是实现数据清洗、建模计算、可视化呈现的核心工具——无论是简单的数据筛选、异常值处理,还是复杂的度量 ...
2026-02-13在互联网运营、产品迭代、用户增长等工作中,“留存率”是衡量产品核心价值、用户粘性的核心指标——而次日留存率,作为留存率体 ...
2026-02-13对CDA(Certified Data Analyst)数据分析师而言,指标是贯穿工作全流程的核心载体,更是连接原始数据与业务洞察的关键桥梁。CDA ...
2026-02-13在机器学习建模实操中,“特征选择”是提升模型性能、简化模型复杂度、解读数据逻辑的核心步骤——而随机森林(Random Forest) ...
2026-02-12在MySQL数据查询实操中,按日期分组统计是高频需求——比如统计每日用户登录量、每日订单量、每日销售额,需要按日期分组展示, ...
2026-02-12对CDA(Certified Data Analyst)数据分析师而言,描述性统计是贯穿实操全流程的核心基础,更是从“原始数据”到“初步洞察”的 ...
2026-02-12备考CDA的小伙伴,专属宠粉福利来啦! 不用拼运气抽奖,不用复杂操作,只要转发CDA真题海报到朋友圈集赞,就能免费抱走实用好礼 ...
2026-02-11在数据科学、机器学习实操中,Anaconda是必备工具——它集成了Python解释器、conda包管理器,能快速搭建独立的虚拟环境,便捷安 ...
2026-02-11在Tableau数据可视化实操中,多表连接是高频操作——无论是将“产品表”与“销量表”连接分析产品销量,还是将“用户表”与“消 ...
2026-02-11在CDA(Certified Data Analyst)数据分析师的实操体系中,统计基本概念是不可或缺的核心根基,更是连接原始数据与业务洞察的关 ...
2026-02-11在数字经济飞速发展的今天,数据已成为核心生产要素,渗透到企业运营、民生服务、科技研发等各个领域。从个人手机里的浏览记录、 ...
2026-02-10在数据分析、实验研究中,我们经常会遇到小样本配对数据的差异检验场景——比如同一组受试者用药前后的指标对比、配对分组的两组 ...
2026-02-10在结构化数据分析领域,透视分析(Pivot Analysis)是CDA(Certified Data Analyst)数据分析师最常用、最高效的核心实操方法之 ...
2026-02-10